We’ve known since 2016 that LSTM networks can be used to generate novel and valid SMILES strings of novel molecules after being trained on a dataset of

We’ve known since 2016 that LSTM networks can be used to generate novel and valid SMILES strings of novel molecules after being trained on a dataset of
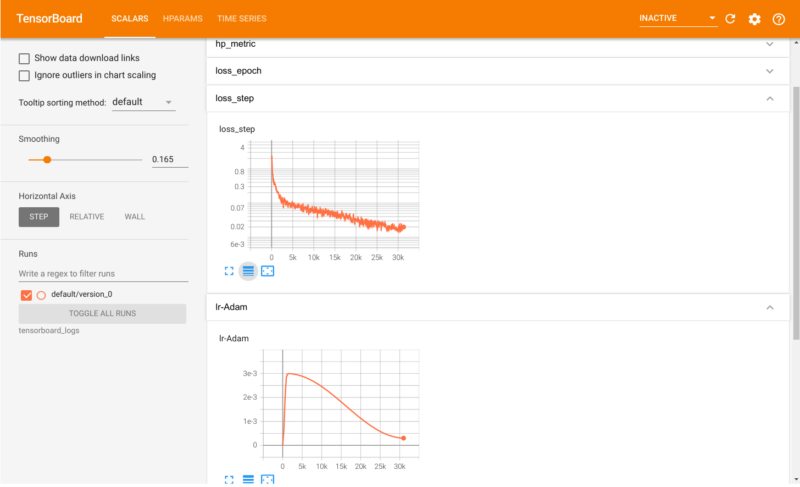
In the last blogpost I covered how LSTM-to-LSTM networks could be used to “translate” reactants into products of chemical reactions. Performance was however not very good of

I have been writing a lot about how to use SMILES together with deep learning architectures such as RNNs and LSTM networks to perform various cheminformatic and
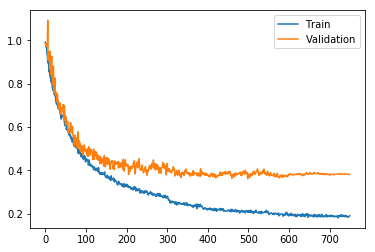
Last blog-post I showed how to use PyTorch to build a feed forward neural network model for molecular property prediction (QSAR: Quantitative structure-activity relationship). RDKit was used
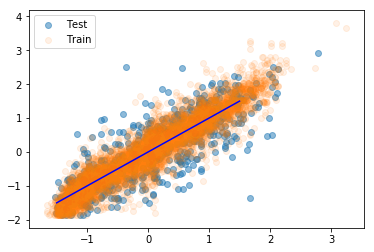
In my previous blogposts I’ve entirely been using Keras for my neural networks. Keras as a stand-alone is now no longer active developed, but are instead now