In the last blogpost I covered how LSTM-to-LSTM networks could be used to “translate” reactants into products of chemical reactions. Performance was however not very good of
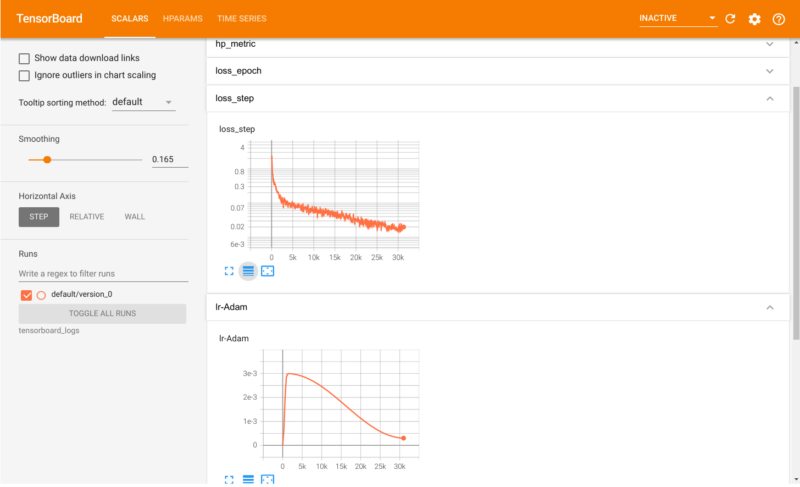

In the last blogpost I covered how LSTM-to-LSTM networks could be used to “translate” reactants into products of chemical reactions. Performance was however not very good of
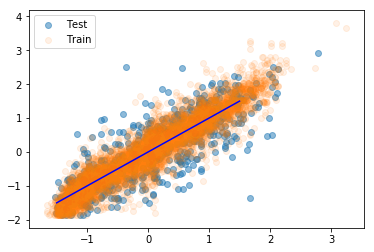
In my previous blogposts I’ve entirely been using Keras for my neural networks. Keras as a stand-alone is now no longer active developed, but are instead now
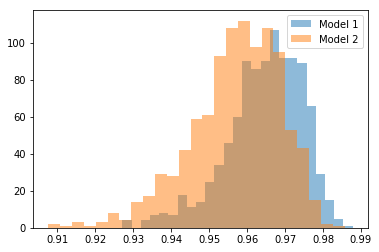
Some time ago I stumbled upon some work by Patrick Walters which shows that correlation coefficients have a rather large standard error when the sample sets sizes
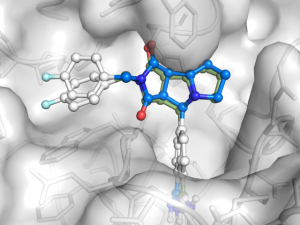
In the two previous blog posts Ligand docking with Smina and Never use re-docking for …, it was demonstrated how easy it is to dock a small ligand

Re-docking of ligands found in PDB files are often used as a fast evaluation of a docking program before working with designed or other ligands. However this
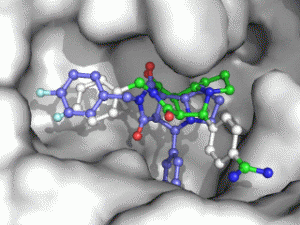
Moleculer Docking is a powerful technique for studying potential ligand-receptor interactions. It can be done with free tools. In this blog post I showcase the docking program