Non-conditional De Novo molecular Generation with Transformer Encoders
We’ve known since 2016 that LSTM networks can be used to generate novel and valid SMILES strings of novel molecules after being trained on a dataset of existing SMILES strings Se previous blog-post. The SMILES strings generated corresponds to molecules with similar properties as the ones the networks were trained on. A lot of techniques has since been made to further control the generation, transfer-learning, reinforcement learning, conditional generation etc.
However cool LSTMs are, they seem to be giving way to the transformer architecture in NLP. But transformers are not only about transforming things, the attention based encoder and decoder blocks have been used in auto-regressive models for text generation such as the GPT style transformers and in bidirectional encoders such as the BERT style transformers. In this blog-post I’ll showcase a simple example of an auto-regressive way the transformer encoder can be used to generate new SMILES strings.
First an import of some of the usual suspects.
%load_ext tensorboard import pandas as pd import numpy as np import matplotlib.pyplot as plt import sys, os sys.version
'3.7.10 (default, Feb 20 2021, 21:17:23) \n[GCC 7.5.0]'
Then we install RDKit via the kora package. It installs RDKit by downloading and unpacking a pre-compiled tar ball, so it’s super fast to do as I’m running on a fresh instance in Google colab.
!pip install kora -q import kora.install.rdkit
[K |████████████████████████████████| 61kB 3.7MB/s
[K |████████████████████████████████| 61kB 7.8MB/s
[?25h
Now that RDKit is installed we can import the needed RDKit modules.
from rdkit import Chem from rdkit.Chem.Draw import IPythonConsole from rdkit.Chem import PandasTools from rdkit.Chem import Descriptors
Import of torch and PyTorch lightning
!pip -q install pytorch-lightning import torch from torch.utils.data import Dataset, DataLoader from pytorch_lightning import LightningDataModule, LightningModule
[K |████████████████████████████████| 849kB 5.1MB/s
[K |████████████████████████████████| 184kB 10.6MB/s
[K |████████████████████████████████| 829kB 14.9MB/s
[K |████████████████████████████████| 112kB 21.6MB/s
[K |████████████████████████████████| 276kB 17.6MB/s
[K |████████████████████████████████| 1.3MB 23.8MB/s
[K |████████████████████████████████| 143kB 35.0MB/s
[K |████████████████████████████████| 296kB 34.2MB/s
[?25h Building wheel for future (setup.py) ... [?25l[?25hdone
Building wheel for PyYAML (setup.py) ... [?25l[?25hdone
Creating the tokenizer
To convert the SMILES strings into tensors for the deep neural network, we’ll code a simple character based tokenizer. Other tokenization schemes are possible as well, such as treating atoms as single tokens (e.g. Cl and Br), but this works reasonable good. For a bigger project that includes on-the-fly SMILES augmentation, take a look at https://github.com/EBjerrum/molvecgen. The molvecgen project is however targeted at Keras and could use an update to provide tensors in the PyTorch way.
# A simple tokenizer
class SimpleTokenizer():
def __init__(self):
self.start = "^"
self.end = "$"
self.unknown = "?"
self.pad = " "
def create_vocabulary(self, smiles):
charset = set("".join(list(smiles)))
#Important that pad gets value 0
self.tokenlist = [self.pad, self.unknown, self.start, self.end] + list(charset)
@property
def tokenlist(self):
return self._tokenlist
@tokenlist.setter
def tokenlist(self, tokenlist):
self._tokenlist = tokenlist
#create the dictionaries
self.char_to_int = {c:i for i,c in enumerate(self._tokenlist)}
self.int_to_char = {i:c for c,i in self.char_to_int.items()}
def vectorize(self, smiles):
return [self.char_to_int[self.start]] + [self.char_to_int.get(char, self.char_to_int[self.unknown]) for char in smiles] + [self.char_to_int[self.end]]
def devectorize(self, tensor):
return [self.int_to_char[i] for i in tensor]
@property
def n_tokens(self):
return len(self.int_to_char)
The dataset just customized how to get the data from the dataframe.
# Build a dataset, that returns tensors
class MolDataset(Dataset):
def __init__(self, data, tokenizer):
self.data = data
self.tokenizer = tokenizer
def __len__(self):
return len(self.data)
def __getitem__(self, idx):
smiles = self.data.smiles.iloc[idx]
tensor = self.tokenizer.vectorize(smiles)
return tensor
Bringing it together in a PyTorch-Lightning datamodule
For the model and training we’ll use PyTorch-lightning as this is a convenient way to organize the data and model into modules. First installation of the PyTorch-lightning package.
To have some SMILES molecules to play with, we will get a tranch from Zinc. Zinc is organized into smaller files in directories based on molecular weight and predicted logP. We just need one that is reasonable fast to download and of an adequate size. Last time I went through how a tokenizer and PyTorch datasets and data-loaders can be integrated into a reusable PyTorch lightning data-module, so I’ll just provide the code here for creating a PyTorch lightning SMILES data-module of a shard from Zinc.
We found that randomizing the atom-order of the molecules and using non-canonical SMILES increased the quality of the generated molecules. So the randomize_smiles_atom_order method has been added to the data-module. Randomizing the atom-order and creating new non-canonical SMILES works as data-augmentation and the dataset size can be increased orders of magnitude in size https://arxiv.org/abs/1703.07076. However, I want to keep this blog-post simple and fast to run through, so I just do it a single time. Even with a single pass the quality of the SMILES generation is improved: https://doi.org/10.1186/s13321-019-0393-0. This is tested with LSTM networks, but I think it’s reasonable to expect it also to increase the quality when using transformers.
class ZincShardDataModule(LightningDataModule):
def __init__(self):
super().__init__()
self.tokenizer = SimpleTokenizer()
self.val_size = 200
self.train_size = 200000
self.batch_size = 128
self.shard = "EEBD" #Shard must be > self.val_size + self.train_size
def prepare_data(self):
# called only on 1 GPU, no assignments of state!
#e.g. use to download_dataset(), tokenize(), build_vocab()
os.system(f"mkdir -pv {self.shard[0:2]} && wget -c http://files.docking.org/2D/{self.shard[0:2]}/{self.shard}.smi -O {self.shard[0:2]}/{self.shard}.smi")
def randomize_smiles_atom_order(self, smiles):
mol = Chem.MolFromSmiles(smiles)
atom_idxs = list(range(mol.GetNumAtoms()))
np.random.shuffle(atom_idxs)
mol = Chem.RenumberAtoms(mol,atom_idxs)
return Chem.MolToSmiles(mol, canonical=False)
def custom_collate_and_pad(self, batch):
#Batch is a list of vectorized smiles
tensors = [torch.tensor(l) for l in batch]
#pad and transpose, pytorch RNNs (and now transformers) expect (sequence,batch, features) batch) dimensions
tensors = torch.nn.utils.rnn.pad_sequence(tensors)
return tensors
def setup(self):
#Load data
self.data = pd.read_csv(f"{self.shard[0:2]}/{self.shard}.smi", sep = " ", nrows = self.val_size + self.train_size)
#Atom order randomize SMILES
self.data["smiles"] = self.data["smiles"].apply(self.randomize_smiles_atom_order)
#Initialize Tokenizer
self.tokenizer.create_vocabulary(self.data.smiles.values)
#Create splits for train/val
np.random.seed(seed=42)
idxs = np.array(range(len(self.data)))
np.random.shuffle(idxs)
val_idxs, train_idxs = idxs[:self.val_size], idxs[self.val_size:self.val_size+self.train_size]
self.train_data = self.data.iloc[train_idxs]
self.val_data = self.data.iloc[val_idxs]
def train_dataloader(self):
dataset = MolDataset(self.train_data, self.tokenizer)
return DataLoader(dataset, batch_size=self.batch_size, pin_memory=True,collate_fn=self.custom_collate_and_pad)
def val_dataloader(self):
dataset = MolDataset(self.val_data, self.tokenizer)
return DataLoader(dataset, batch_size=self.batch_size, pin_memory=True,collate_fn=self.custom_collate_and_pad, shuffle=False)
It should now be ready to download and prepare the data for training.
mydm = ZincShardDataModule() mydm.prepare_data() mydm.setup()
mydm.data.head()
| smiles | zinc_id | |
|---|---|---|
| 0 | c1(Oc2ccc(OC)cc2)ccc([N+](=O)[O-])cc1C(NCC(=O)… | ZINC000000132042 |
| 1 | c12cc(NS(c3ccc([N+]([O-])=O)cc3)(=O)=O)ccc1COC2=O | ZINC000000178081 |
| 2 | N(n1c(=O)c2ccccc2nc1)C(c1cc([N+]([O-])=O)c(Cl)… | ZINC000000183321 |
| 3 | O=C(Nn1c(=O)c2ccccc2nc1)c1cc(Cl)ccc1[N+](=O)[O-] | ZINC000000183346 |
| 4 | C(=C/c1ccc([N+]([O-])=O)cc1)\C(=O)Nn1c(=O)c2cc… | ZINC000000183354 |
A quick check that the dataloaders provide tensors.
dl = mydm.val_dataloader() test_batch = next(iter(dl)) test_batch
tensor([[ 2, 2, 2, ..., 2, 2, 2],
[28, 6, 16, ..., 28, 16, 28],
[15, 15, 15, ..., 15, 19, 15],
...,
[ 0, 0, 0, ..., 0, 0, 0],
[ 0, 0, 0, ..., 0, 0, 0],
[ 0, 0, 0, ..., 0, 0, 0]])
plt.matshow(test_batch.T)

Properties of the dataset
Let’s quickly test how the molecular weight distribution of the dataset looks like.
PandasTools.AddMoleculeColumnToFrame(mydm.data,'smiles','ROMol') mydm.data["MW"] = mydm.data.ROMol.apply(Descriptors.MolWt) _ = plt.hist(mydm.data["MW"])

This is a relatively narrow distribution with clear cutoffs. It’ll be interesting to see how close the distribution of the generated molecules will be.
Coding the PyTorch-Lightning model
Next up is the creation of the transformer model. For some reason PyTorch has transformer models, transformer layers and such, but not the positional encoding that was used in the original paper. It is however in one of the examples, so we’ll grab the function from there. Basically it creates a positional encoding and adds it to the tensor, so that the transformer model can figure out the distance and order of the tokens.
!wget -c https://raw.githubusercontent.com/pytorch/examples/master/word_language_model/model.py from model import PositionalEncoding
--2021-05-01 10:51:49-- https://raw.githubusercontent.com/pytorch/examples/master/word_language_model/model.py
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.108.133, 185.199.109.133, 185.199.110.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.108.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 6367 (6.2K)
Saving to: ‘model.py’ model.py 100%[===================>] 6.22K --.-KB/s in 0s 2021-05-01 10:51:49 (62.5 MB/s) - ‘model.py’ saved [6367/6367]
Now we have all we need to create our SMILES generator model as a PyTorch lightning module. PyTorch lightning again expects some standard functions for the trainer to work, namely for getting and optimizer object, and to take a step (for both train and val). Setup of the layers , the forward function and the shared steps are just for our own convenience. The init function runs the save_hyperparameters() function, which saves all the arguments passed at init into a hparams object on the model. This way they will also latter get logged :-). The setup_layers functions creates the layers that we need for the model.
First the embedding layer that converts the tensors of integers into tensors of embedding vectors of the needed dimension (d_model). These are then passed on to the positional encoding object, that adds this extra information. A triangular mask is needed to prevent the model from looking “into the future” on the character that it is supposed to predict in an auto-regressive manner. The encoder is created from the encoder_layer and the layer normalization. Lastly is a fully connected linear layer that converts the vectors (d_model size) into the size of the vocabulary (here 36).
The step function is used by the PyTorch lightning training_ and validation_step functions. A key element in these functions here is the shift between the input and output. The input contains the start token, but not the last token, whereas the output skips the start token and has the last character. This way the first character of the SMILES will be the predicted output that is aligned with the start token. The second position will “see” the start token and the first character of the SMILES and will predict the second character of the SMILES and so forth due to triangular masking that is applied. Also note the self.log calls in the step functions. Here we control what information we want to get logged and when (per step or per epoch).
class SmilesEncoderModel(LightningModule):
def __init__(
self,
n_tokens,
d_model=256,
nhead=8,
num_encoder_layers=4,
dim_feedforward=1024,
dropout=0.1,
activation="relu",
max_length = 1000,
max_lr = 1e-3,
epochs = 50,
):
super().__init__()
self.save_hyperparameters() #Populates self.hparams from __init__ options.
self.setup_layers()
def configure_optimizers(self):
optimizer = torch.optim.Adam(self.parameters())
scheduler = torch.optim.lr_scheduler.OneCycleLR(optimizer, max_lr=self.hparams.max_lr,
total_steps=None, epochs=self.hparams.epochs, steps_per_epoch=len(self.train_dataloader()),#We call train_dataloader, just to get the length, is this necessary?
pct_start=6/self.hparams.epochs, anneal_strategy='cos', cycle_momentum=True,
base_momentum=0.85, max_momentum=0.95,
div_factor=1e3, final_div_factor=1e3, last_epoch=-1)
scheduler = {"scheduler": scheduler, "interval" : "step" }
return [optimizer], [scheduler]
def setup_layers(self):
self.embedding = torch.nn.Embedding(self.hparams.n_tokens, self.hparams.d_model)
self.positional_encoder = PositionalEncoding(self.hparams.d_model, dropout=self.hparams.dropout)
encoder_layer = torch.nn.TransformerEncoderLayer(self.hparams.d_model, self.hparams.nhead, self.hparams.dim_feedforward, self.hparams.dropout, self.hparams.activation)
encoder_norm = torch.nn.LayerNorm(self.hparams.d_model)
self.encoder = torch.nn.TransformerEncoder(encoder_layer, self.hparams.num_encoder_layers, encoder_norm)
self.fc_out = torch.nn.Linear(self.hparams.d_model, self.hparams.n_tokens)
def _generate_square_subsequent_mask(self, sz):
mask = (torch.triu(torch.ones(sz, sz)) == 1).transpose(0, 1)
mask = mask.float().masked_fill(mask == 0, float('-inf')).masked_fill(mask == 1, float(0.0))
return mask
def forward(self, features):
mask = self._generate_square_subsequent_mask(features.shape[0]).to(self.device)
embedded = self.embedding(features)
positional_encoded = self.positional_encoder(embedded)
encoded = self.encoder(positional_encoded, mask=mask)
out_2= self.fc_out(encoded)
return out_2
def step(self, batch):
batch = batch.to(self.device)
prediction = self.forward(batch[:-1])#Skipping the last char
loss = torch.nn.functional.cross_entropy(prediction.transpose(0,1).transpose(1,2), batch[1:].transpose(0,1))#Skipping the first char
return loss
def training_step(self, batch, batch_idx):
self.train()
loss = self.step(batch)
self.log('loss', loss, on_step=True, on_epoch=True, prog_bar=True, logger=True)
return loss
def validation_step(self, batch, batch_idx):
self.eval()
loss = self.step(batch)
self.log('val_loss', loss, on_step=False, on_epoch=True, prog_bar=True, logger=True)
return loss
With that in place we can instantiate our PyTorch lightning model. I keep the dimensions low with two layers and d_model of 256 so that training is fast for demonstration purposes. Larger models (e.g. 4-layers) are more the default when it comes to transformers used for chemistry.
mydm.batch_size = 256
model = SmilesEncoderModel(n_tokens=mydm.tokenizer.n_tokens, num_encoder_layers=4, d_model=512, nhead=8, epochs=25, max_lr=8e-4)
Let’s just make a sanity check by passing our test_batch from before through the model.
out = model.forward(test_batch) out.size()
torch.Size([79, 128, 37])
plt.matshow(out[:,0,:].detach())

Training the model
Next is just to decide that we want also to log the learning rate and that we want to use the TensorBoard logger when we create the trainer object.
import pytorch_lightning
lr_logger =pytorch_lightning.callbacks.lr_monitor.LearningRateMonitor(logging_interval="epoch")
tb_logger = pytorch_lightning.loggers.TensorBoardLogger('tensorboard_logs/')
trainer = pytorch_lightning.Trainer(
logger=tb_logger,
callbacks=[lr_logger],
max_epochs=model.hparams.epochs,
gpus=1,
progress_bar_refresh_rate=20)
GPU available: True, used: True
TPU available: False, using: 0 TPU cores
The TensorBoard extension for Google colab is quite convenient to monitor the training interactively, and also for keeping track of different models performance and hyper parameters.
%tensorboard --logdir tensorboard_logs
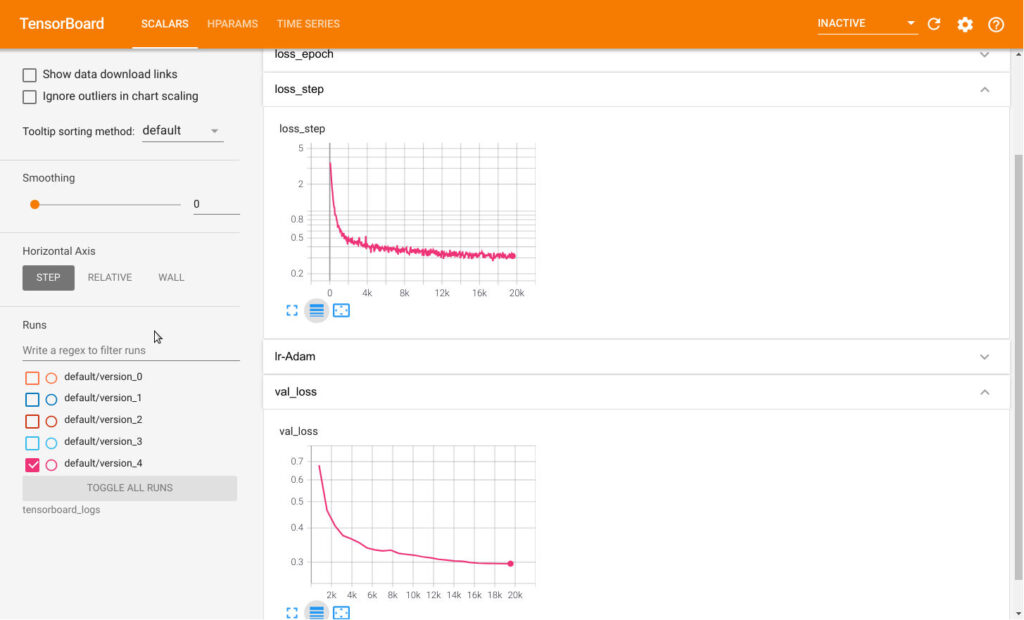 Now we can start training using the trainer object. Enjoy. I always finds it facinating to watch the training loss drop as training progresses.
Now we can start training using the trainer object. Enjoy. I always finds it facinating to watch the training loss drop as training progresses.
trainer.fit(model, mydm)
 Saving a little bundle with the model, the hyperparameters and the tokenlist from the tokenizer can come in handy later.
Saving a little bundle with the model, the hyperparameters and the tokenlist from the tokenizer can come in handy later.
import pickle
save_dict = {"state_dict": model.state_dict(), "tokenlist": mydm.tokenizer.tokenlist, "hparams":model.hparams}
pickle.dump(save_dict, open("drive/MyDrive/temp/deNovoFormer_25.pickle","wb"))
Sampling the model
Now that we have trained the model, we can see how the test_batch now looks. It looks a lot less random than before and we note that there’s some padding in the end (position 0) sampled with high probability, possibly after the end token.
model.eval() out = model.forward(test_batch) plt.matshow(out[:,7,:].detach())

In order to sample a SMILES string, we need to write a small sample loop. The sampler creates an empty tensor and seeds this with the start token. Then this is passed into the model and the probability distribution used to sample the next character which are then added to the tensor, before it’s passes into the model again. Only the slice of the tensor needed for prediction of the next character is passes into the model to save computations. If the stop token is sampled, the iteration stops. This is different from the LSTM based sampling, where simply the hidden states are passed along from prediction to prediction. Here, a longer and longer tensor needs to be passed into the model, which contributes to making the sampling slower.
class Sampler():
def __init__(self, model, tokenizer):
self.model = model
self.tokenizer = tokenizer
self.max_len = 128
def sample(self):
model.eval()
sample_tensor = torch.zeros((self.max_len,1), dtype=torch.long).to(model.device)
sample_tensor[0,0] = self.tokenizer.char_to_int[self.tokenizer.start]
with torch.no_grad():
for i in range(self.max_len-1):
tensor = sample_tensor[:i+1]
logits = self.model.forward(tensor)[-1]
probabilities = torch.nn.functional.softmax(logits, dim=1).squeeze()
sampled_char = torch.multinomial(probabilities,1)
sample_tensor[i+1,0] = sampled_char
if sampled_char == self.tokenizer.char_to_int[self.tokenizer.end]:
#print("Done")
break
smiles = "".join(self.tokenizer.devectorize(sample_tensor.squeeze().detach().cpu().numpy())).strip("^$ ")
return smiles
def sample_multi(self, n):
smiles_list = []
for i in range(n):
smiles_list.append(self.sample())
return smiles_list
Moving the model to the GPU helps on inference speed
model.to("cuda")
model.device
device(type='cuda', index=0)
sampler = Sampler(model, mydm.tokenizer) smiles_list = sampler.sample_multi(100)
Sampling would be faster if we did it in batches, but to keep things simple, it’s done one-by-one in this example Sampler class.
We’ll parse the generated SMILES strings into RDKit molecules and compute the fraction of unparsable SMILES strings.
parsed_mols = np.array([Chem.MolFromSmiles(s) for s in smiles_list])
RDKit ERROR: [16:31:59] Can't kekulize mol. Unkekulized atoms: 16 17 21
RDKit ERROR:
RDKit ERROR: [16:31:59] Can't kekulize mol. Unkekulized atoms: 0 4 5 6 19 20 22 23 24
RDKit ERROR:
RDKit ERROR: [16:31:59] Can't kekulize mol. Unkekulized atoms: 0 2 18 21 23
RDKit ERROR:
print("Percentage invalid")
(parsed_mols == None).sum()/len(parsed_mols)
Percentage invalid
0.03
The model has definitely learned something about creation SMILES strings character by character, but there may be room for improvement. SMILES augmentation, larger datasets, larger models and longer training would likely improve the results even more.
We can take a look at the molecules
parsed_mols[1]
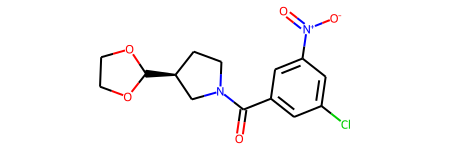
They don’t look that weird to me.
How does the distribution of the molecular weights looks?
_ = plt.hist(list(map(Descriptors.MolWt, parsed_mols[parsed_mols != None]))) plt.axvline(x=min(mydm.data["MW"]), c="r") plt.axvline(x=max(mydm.data["MW"]), c="r")

The molecular weight of the generated molecules are both smaller and larger than in the training set. The distribution of the de novo generated molecules are however centered on the previous distribution.
Are the molecules novel? We’ll create sets of InChi keys and see of there’s overlap.
denovo_inchi = set(map(Chem.inchi.MolToInchi, parsed_mols[parsed_mols != None])) train_inchi = set(map(Chem.inchi.MolToInchi, mydm.data.ROMol))
[1;30;43mStreaming output truncated to the last 5000 lines.[0m
RDKit WARNING: [16:35:19] WARNING: Charges were rearranged
RDKit WARNING: [16:35:19] WARNING: Charges were rearranged
...
This produces a fair set of warning about charges. I think ZINC has charged molecules ready for docking, maybe this is what triggers the warnings. Or perhaps the charge separated representation for nitro groups.
denovo_inchi.intersection(train_inchi)
{'InChI=1S/C12H17BrN4O2/c1-15(2)7-9-3-4-16(8-9)12-11(13)5-10(6-14-12)17(18)19/h5-6,9H,3-4,7-8H2,1-2H3/t9-/m1/s1',
'InChI=1S/C13H18N2O4S2/c1-10-7-12(15(16)17)4-5-13(10)21(18,19)14-8-11-3-2-6-20-9-11/h4-5,7,11,14H,2-3,6,8-9H2,1H3/t11-/m1/s1',
'InChI=1S/C13H19N3O4S2/c1-8-5-9(7-15(8)10-3-4-10)14-13-11(16(17)18)6-12(21-13)22(2,19)20/h6,8-10,14H,3-5,7H2,1-2H3/t8-,9-/m0/s1',
'InChI=1S/C14H20N2O5S/c1-11-6-7-13(9-14(11)16(17)18)22(19,20)15(2)10-12-5-3-4-8-21-12/h6-7,9,12H,3-5,8,10H2,1-2H3/t12-/m0/s1',
'InChI=1S/C16H16N2O5S/c1-11-3-5-12(6-4-11)10-17-16(19)13-7-8-15(24(2,22)23)14(9-13)18(20)21/h3-9H,10H2,1-2H3,(H,17,19)',
'InChI=1S/C16H18N4O4/c1-11(21)19-8-6-12(7-9-19)16-17-15(18-24-16)10-13-4-2-3-5-14(13)20(22)23/h2-5,12H,6-10H2,1H3',
'InChI=1S/C16H21FN2O5/c1-4-24-13-8-16(9-20,15(13,2)3)18-14(21)11-6-5-10(19(22)23)7-12(11)17/h5-7,13,20H,4,8-9H2,1-3H3,(H,18,21)/t13-,16+/m0/s1',
'InChI=1S/C17H20N4O4/c1-12-11-25-8-6-13(12)10-18-17(22)16-5-7-20(19-16)14-3-2-4-15(9-14)21(23)24/h2-5,7,9,12-13H,6,8,10-11H2,1H3,(H,18,22)/t12-,13+/m0/s1',
'InChI=1S/C17H25N3O4/c1-13(12-21)5-4-8-18-17(22)14-6-7-15(16(11-14)20(23)24)19-9-2-3-10-19/h6-7,11,13,21H,2-5,8-10,12H2,1H3,(H,18,22)/t13-/m0/s1',
'InChI=1S/C18H20N2O4/c1-13-7-8-16(20(23)24)10-17(13)18(22)19-11-15(12-21)9-14-5-3-2-4-6-14/h2-8,10,15,21H,9,11-12H2,1H3,(H,19,22)/t15-/m1/s1'}
Apparently some of the generated molecules belonged to the train set, so the network has started to learn the dataset by hearth. But still a lot of novel molecules in there.
So I hope this walk-through of using transformers models was instructive. They can certainly be used for de novo generation of molecules, but if they are better than LSTM networks for this particular task would require more careful comparison by keeping the datasets fixed and doing proper hyperparameter tuning for both model types. I think they are a bit more complicated to work with than LSTM networks. However, as we saw in a previous blog-post, LSTM networks fails quite miserably when it comes to molecular transformation tasks such as reaction prediction, and I hope to soon get the time to write a blog-post about implementing a transformer model for reaction prediction.
Pingback: Cambridge Cheminformatics Newsletter, 20 May 2021 – DrugDiscovery.NET – AI in Drug Discovery
What a fantastic presentation!
I am wondering if we can still use the dataset from your QSAR model which is SLC6A4_active_excape_export.csv. Then we use the PCX50 from the dataset as the Y variable and compute the X variable using output from this transformer model with smiles string as the input for a regression model. analysis, what kind of coding change is needed? Is it possible to do that?
Yes, it would be entirely doable using transformers to build QSAR models from e.g. the SC6A4 dataset or any other molecular activity dataset. Usually there is put in a pooling layer on the output from the transformer to then feed into a standard dense network for the final prediction, or alternatively take a single output vector aligned with a token. In my experience it doesn’t work so great from scratch, so some form of pretraining is advised. The MolBert model seem to get nice property prediction results: https://arxiv.org/abs/2011.13230
When I run trainer,fit(), I got the following error.
: TypeError: setup() got an unexpected keyword argument ‘stage’
6 #model = DataModule(n_tokens = 35, num_encoder_layers=2, d_model=512, nhead=8, epochs=25, max_lr=8e-4)
7
—-> 8 trainer.fit(model, mydm)
D:\users\envs\pytorch\lib\site-packages\pytorch_lightning\trainer\trainer.py in fit(self, model, train_dataloaders, val_dataloaders, datamodule, train_dataloader, ckpt_path)
734 train_dataloaders = train_dataloader
–> 735 self._call_and_handle_interrupt(
736 self._fit_impl, model, train_dataloaders, val_dataloaders, datamodule, ckpt_path
737 )
D:\users\envs\pytorch\lib\site-packages\pytorch_lightning\trainer\trainer.py in _call_and_handle_interrupt(self, trainer_fn, *args, **kwargs)
680 “””
681 try:
–> 682 return trainer_fn(*args, **kwargs)
683 # TODO: treat KeyboardInterrupt as BaseException (delete the code below) in v1.7
684 except KeyboardInterrupt as exception:
D:\users\envs\pytorch\lib\site-packages\pytorch_lightning\trainer\trainer.py in _fit_impl(self, model, train_dataloaders, val_dataloaders, datamodule, ckpt_path)
768 # TODO: ckpt_path only in v1.7
769 ckpt_path = ckpt_path or self.resume_from_checkpoint
–> 770 self._run(model, ckpt_path=ckpt_path)
771
772 assert self.state.stopped
D:\users\envs\pytorch\lib\site-packages\pytorch_lightning\trainer\trainer.py in _run(self, model, ckpt_path)
1130 self.call_hook(“on_before_accelerator_backend_setup”)
1131 self.accelerator.setup_environment()
-> 1132 self._call_setup_hook() # allow user to setup lightning_module in accelerator environment
1133
1134 # check if we should delay restoring checkpoint till later
D:\users\envs\pytorch\lib\site-packages\pytorch_lightning\trainer\trainer.py in _call_setup_hook(self)
1430 if self.datamodule is not None:
1431 self.datamodule.setup(stage=fn)
-> 1432 self.call_hook(“setup”, stage=fn)
1433
1434 self.training_type_plugin.barrier(“post_setup”)
D:\users\envs\pytorch\lib\site-packages\pytorch_lightning\trainer\trainer.py in call_hook(self, hook_name, pl_module, *args, **kwargs)
1481 model_fx = getattr(pl_module, hook_name, None)
1482 if callable(model_fx):
-> 1483 output = model_fx(*args, **kwargs)
1484
1485 # *Bad code alert*
TypeError: setup() got an unexpected keyword argument ‘stage’
Thanks for the interest and for reaching out. I’ve never seen this error before, and as is evident from the stack trace it appears from within the pytorch-lightning framework. It could thus derive from a difference in versions. However, something strange seem to be happening on your line 6, where there is an out commented line: “model = DataModule(n_tokens = 35, num_encoder_layers=2, d_model=512, nhead=8, epochs=25, max_lr=8e-4)”, which is not from my blog-post, hinting that you are not running the code as-is. It would be very strange to try and train a model subclassed from a pytorch DataModule. So unless you reveal your total code (e.g. github), I can’t help you more than this.
I combined the Class SmileEncoderModule with Class DataModule in a single file and the Trainer.fit was able to run under pytorch lightning after initially met with errors.
If we use the dataset from your blog under QSAR model and run the nonconditional denovo molecular generation transformer with PCX50 as the labels in regression model, what sorts of modifications are needed to achieve meaningful outcome? I will greatly appreciate your feedback. Many thanks for your wonderful contribution.
Well, you would need to make several changes. First is to the dataloader, which should return a tuple of a list of vectorized SMILES and a list of the pXC50 values for the mini-batches. Then you would need to change the flow through the network. As an example take the memory and index into the position of e.g. the start-token or a special token you design as a “predict here” token, and feed that into a feed forward neural network that ends in a single neuron with linear activation function as you want regression. than an MSE loss would be fitting. Alternative architecture is to add a pooling layer to the memory (along the sequence dimension), and then feed the pooled vector into the feed forward neural network. You would probably need to device some kind of unsupervised pretraining to get good results.
In your previous article, you have used MolVecGen to vectorize the smiles string for LSTM research, why don’t you use it in place of the simple tokenizer that is used in this article?
Molvecgen only supports one-hot encoding and not the int vector + embedding layer I use here (and I don’t have time to update it). Instead, we have “recently” released PySMILESUtils that supports both: https://github.com/MolecularAI/pysmilesutils. Besides, it can be illustrative with a bare-bones vectorizer.
I assume that pytorch nn transformer module you used here is different from Bert transformer. which has CLS, SEP, tokens id etc. What are the advantages of your choice over others?
Thank you again.
I’m not sure I understand your question. Bert is a different model that is trained for a different purpose. If I would need cls and sep tokens they should be added to the tokenization, not to the neural network modules.
trainer.fit(model, mydm)
I have used the line above and got error: `train_dataloader` must be implemented to be used with the Lightning Trainer
That is strange, since train_dataloader IS defined in the datamodule:
def train_dataloader(self):
dataset = MolDataset(self.train_data, self.tokenizer)
return DataLoader(dataset, batch_size=self.batch_size, pin_memory=True,collate_fn=self.custom_collate_and_pad)
Can you try and call dl = mydm.train_dataloader() and examine what is dl?