Cheminformatics in Excel: linking RDKit with Xlwings

Excel is widely used in businesses all over the world and can be used for many diverse tasks due to the flexibility of the program. I’ve been doing a fair deal of Excel templates over the years to solve an array of very different tasks. I’ve also been using RDKit for a lot of cheminformatics related tasks, so why not combine these tools?
It sounds like a useful idea. There are several options for how it can be done, and I’ll explore one of them in this blog post. One way of linking Excel to Python is via visual basic (VBA), and lucklily this has already been developed in the xlwings framework (https://www.xlwings.org/), which is an open source, BSD-licensed Python library that can make it easier to link excel to python code, either via Macros or user defined functions. It can also be used to script Excel from python, which I normaly do with COM automation via win32com Python module. An alternative proprietary program is PyXLL, https://www.enthought.com/products/pyxll/, which is probably more mature, but I haven’t tested that.
It is fast to install xlwings via pip
pip install xlwings
On my installation I also needed Pillow, The prerequisites can be found at the installation guide: http://docs.xlwings.org/en/stable/addin.html#installation
pip install Pillow
Then the necessary xlwings Excel add-in must be installed. The fastest way is via the provided setup script
xlwings addin install
The next step is to generate an Excel file with the necessary VBA. This is done from the command line with a setup script
xlwings quickstart my_test_project
This will create a directory named my_test_project with two files: a my_test_project.py file and a macro enabled Excel sheet my_test_project.xlsm.
The my_test_project.py is where we will define the Python functions, and the Excel workbook have been set up with the necessary VBA functions to allow xlwings to work.
The my_test_project.py file contains two examples of how the python functions can be called. The first is a regular function, that calls backs to the workbook where it has been called from and writes “hello Xlwings!” in cell A1 on the first worksheet. This must be called from within VBA scripts, and the workbook contains a macro SampleCall() that will execute the function.
import xlwings as xw
def hello_xlwings():
wb = xw.Book.caller()
wb.sheets[0].range("A1").value = "Hello xlwings!"
@xw.func
def hello(name):
return "hello {0}".format(name)
The second one is an example of a user defined function(UDF). It has been decorated with @xw.func which notifies xlwings that this function should be made callable as a UDF. If we open the my_test_project.xlsm and enables macroes and type =hello(“world!”) into a cell, nothing happens. We first have to import the defined functions into the VBA embedded into the workbook. This is done with the “import functions” under the xlwings entry on the ribbon. After doing that, the formula will work as intended (press F9 to refresh). Pressing the “import function” will update the embedded VBA with the functions from the Python file:
Function hello(name)
If TypeOf Application.Caller Is Range Then On Error GoTo failed
hello = Py.CallUDF("my_test_project", "hello", Array(name), ThisWorkbook, Application.Caller)
Exit Function
failed:
hello = Err.Description
End Function
So to make RDKit available we can add a test function. First some import statements
import rdkit from rdkit import Chem from rdkit.Chem import Descriptors, Draw
Then the decorated python functions
@xw.func
def rdkit_version():
return rdkit.__version__
After a refresh of the functions using the “import functions” as before the excel formula =rdkit_version() returns 2016.03.1. which show that it works! Its easy to extend with something more useful, where we calculate some properties of some entered SMILES strings
@xw.func
def rdkit_mol_GetNumAtoms(smiles):
mol = Chem.MolFromSmiles(smiles)
if mol != None:
numatoms = mol.GetNumAtoms()
return numatoms
else:
return 0
@xw.func
def rdkit_LogP(smiles):
mol = Chem.MolFromSmiles(smiles)
if mol != None:
return Descriptors.MolLogP(mol)
else:
return None
Then the functions rdkit_LogP and rdkit_mol_GetNumAtoms can be used to calculate the LogP and number of atoms from SMILES string. It can be generalized even more:
@xw.func
def rdkit_Descriptor(smiles, fname='MolLogP'):
myfunction = getattr(Descriptors, fname)
mol = Chem.MolFromSmiles(smiles)
if mol != None:
return myfunction(mol)
else:
return None
This function allows one to call an arbitrary function from the descriptors module by specifying its name as the second option. =rdkit_Descriptor(“COCCCN”; “NumHAcceptors”) will calculate the number of hydrogen bond acceptors. A list of functions in the descriptors module can be found in the documentation, http://www.rdkit.org/Python_Docs/rdkit.Chem.Descriptors-module.html, or can be generated with the code:
for desc in Descriptors.descList: print desc[0]
To make an interactive descriptor calculator, the list could be pasted into a hidden sheet, referenced as a list with data validation to generate a drop down menu, and the content of the cell referenced by the UDF function.
This way its easy to build an Excel sheet which takes compounds and SMILES, calculates some properties using the general formula and as an example flag them using the Lipinski’s rule of five.
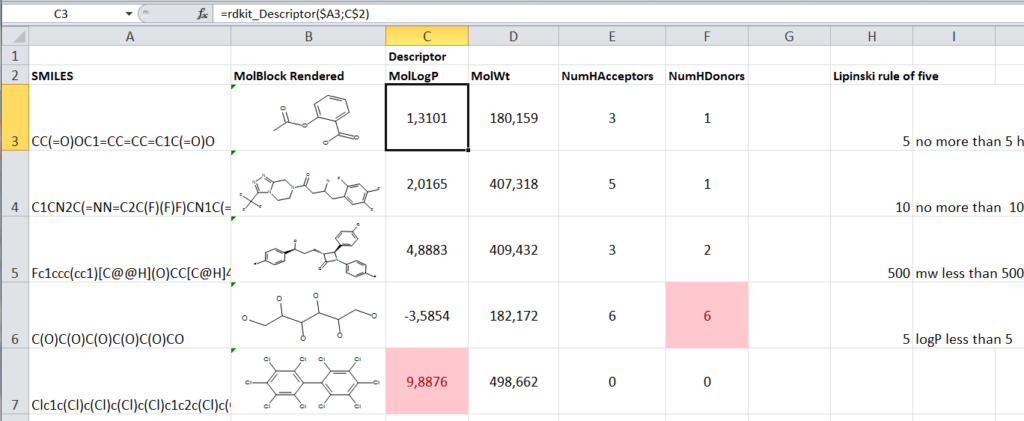
However, having python and all the extensions available for Excel opens up a lot of interesting possibilities, which are not just limited to using Rdkit.
Unfortunately there’s an issue with the xlwings code, which limits the return string to 256 characters. https://github.com/ZoomerAnalytics/xlwings/issues/456
This prevents conversion of SMILES to molfile text with subsequent visualization as could be done with Proteax for Excel so the mol renderings have been made with other means (rdkit4excel + proteax).
We have also seen issues with sharing of workbooks, as the addin, VBA and the python code needs to be in sync, and this have been an issue in deployment.
I’ll probably cover use the rdkit4excel program (https://github.com/janholstjensen/rdkit4excel) in a future blog post. Here we employed the python code directly as a COM automation object which can be used as a plugin for Excel.
Hi there!
Loved your post. Is there any way to show the chemical structures in Excel?
Thank you!
Thanks. I don’t know of an open-source or free solution, but Proteax from Biochemfusion can do it. http://www.biochemfusion.com/products/proteax_for_spreadsheets/