In the last blogpost I covered how LSTM-to-LSTM networks could be used to “translate” reactants into products of chemical reactions. Performance was however not very good of
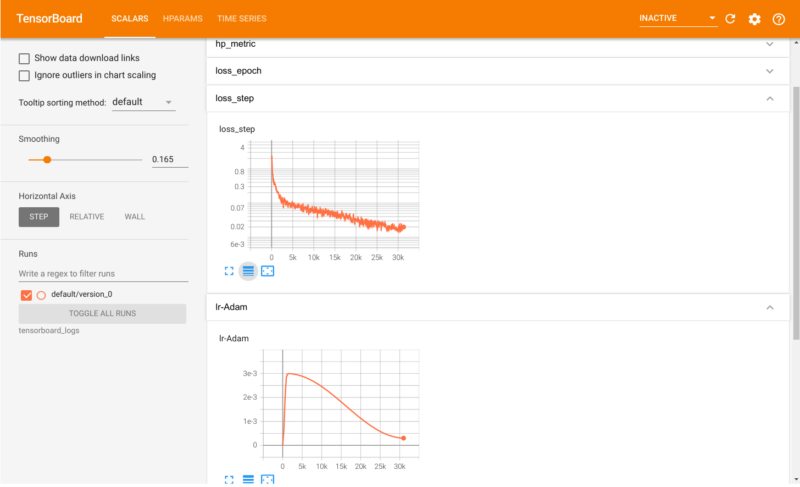

In the last blogpost I covered how LSTM-to-LSTM networks could be used to “translate” reactants into products of chemical reactions. Performance was however not very good of
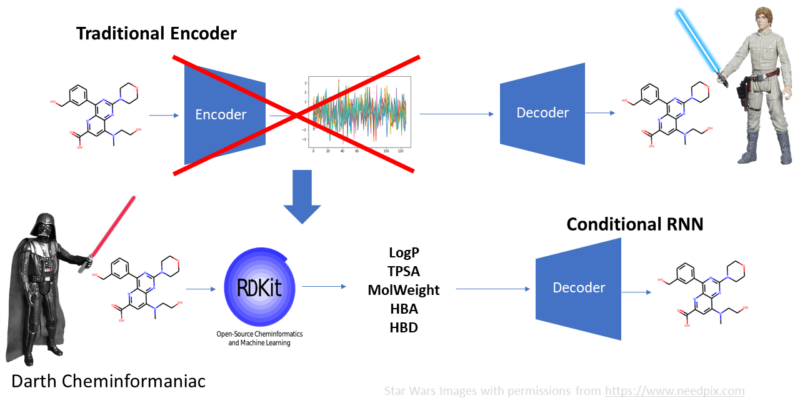
Long time ago in a GPU far-far away, the deep learning rebels are happy. They have created new ways of working with chemistry using deep learning technology
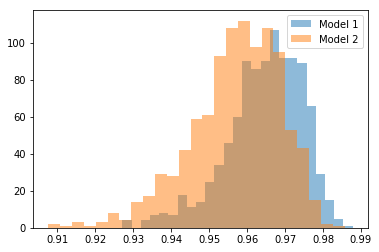
Some time ago I stumbled upon some work by Patrick Walters which shows that correlation coefficients have a rather large standard error when the sample sets sizes

Earlier I wrote a blog post about how to build SMILES based autoencoders in Keras. It has since been a much visited page, so the topic seems

During my postdoc project at the Chemometrics and Analytical Technology section at Copenhagen University I worked with modeling of spectroscopical data with PLS models. Chemometrics is “the

UPDATE: Be sure to check out the follow-up to this post if you want to improve the model: Learn how to improve SMILES based molecular autoencoders with

The film Inception with Leonardo Di Caprio is about dreams in dreams, and gave rise to the meme “We need to go deeper”. The title has also

The process of expanding an otherwise limited dataset in order to more efficiently train a neural network is known as Data Augmentation For images there have been

Neural Networks are interesting algorithms, but sometimes also a bit spooky. In this blog post I explore the possibilities for teaching the neural networks to generate completely
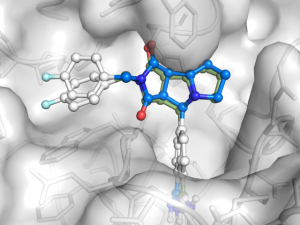
In the two previous blog posts Ligand docking with Smina and Never use re-docking for …, it was demonstrated how easy it is to dock a small ligand