Last blog-post I showed how to use PyTorch to build a feed forward neural network model for molecular property prediction (QSAR: Quantitative structure-activity relationship). RDKit was used
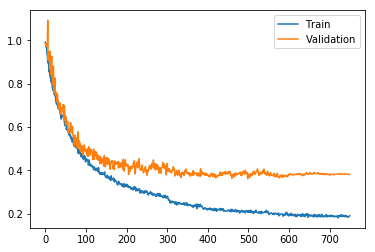

Last blog-post I showed how to use PyTorch to build a feed forward neural network model for molecular property prediction (QSAR: Quantitative structure-activity relationship). RDKit was used
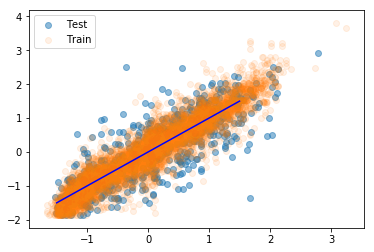
In my previous blogposts I’ve entirely been using Keras for my neural networks. Keras as a stand-alone is now no longer active developed, but are instead now
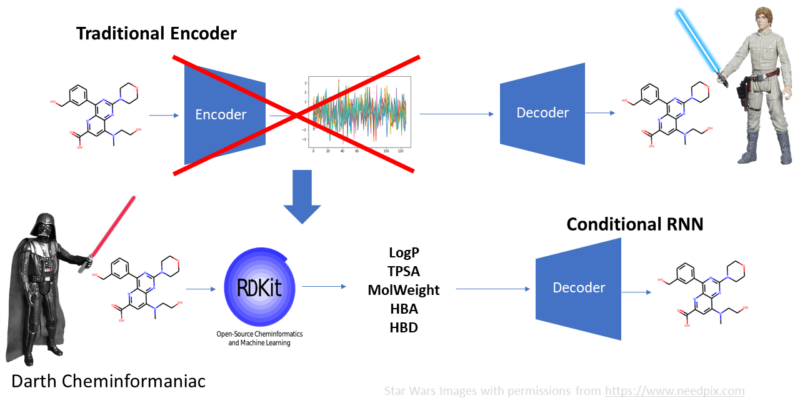
Long time ago in a GPU far-far away, the deep learning rebels are happy. They have created new ways of working with chemistry using deep learning technology
Not so long ago Greg Landrum published a blog post with an example of how the SVG rendering from RDKit in a jupyter notebook can be
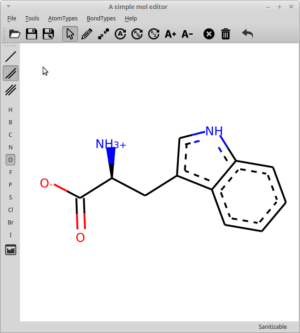
At the RDKit UGM 2018 in Cambridge I made a lightning talk where I show cased rdEditor. I’ve wanted to write a bit about it for some
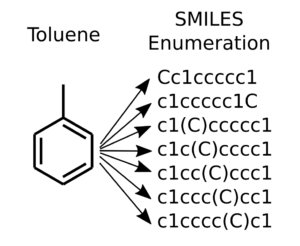
The SMILES enumeration code at GitHub has been revamped and revised into an object for easier use. It can work in conjunction with a SMILES iterator object

The film Inception with Leonardo Di Caprio is about dreams in dreams, and gave rise to the meme “We need to go deeper”. The title has also

Excel is widely used in businesses all over the world and can be used for many diverse tasks due to the flexibility of the program. I’ve been
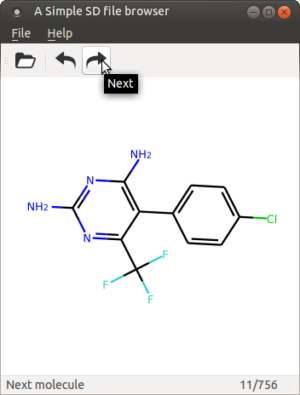
One of the more popular blog post based on monthly visitors is the old Create a Simple Object Oriented GUIDE GUI in MatLAB, but since I don’t

The process of expanding an otherwise limited dataset in order to more efficiently train a neural network is known as Data Augmentation For images there have been