We’ve known since 2016 that LSTM networks can be used to generate novel and valid SMILES strings of novel molecules after being trained on a dataset of

We’ve known since 2016 that LSTM networks can be used to generate novel and valid SMILES strings of novel molecules after being trained on a dataset of
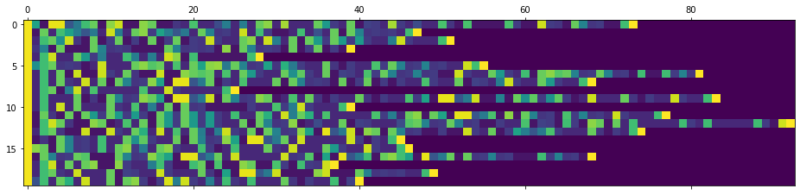
In this blogpost I’ll show how to predict chemical reactions with a sequence to sequence network based on LSTM cells. It’s the same principle as IBM’s RXN
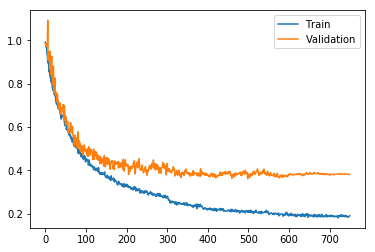
Last blog-post I showed how to use PyTorch to build a feed forward neural network model for molecular property prediction (QSAR: Quantitative structure-activity relationship). RDKit was used

Earlier I wrote a blog post about how to build SMILES based autoencoders in Keras. It has since been a much visited page, so the topic seems
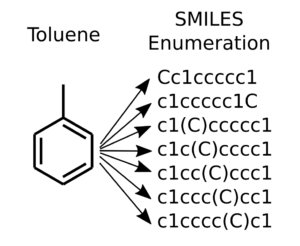
The SMILES enumeration code at GitHub has been revamped and revised into an object for easier use. It can work in conjunction with a SMILES iterator object