Neural Networks are interesting algorithms, but sometimes also a bit spooky. In this blog post I explore the possibilities for teaching the neural networks to generate completely


Neural Networks are interesting algorithms, but sometimes also a bit spooky. In this blog post I explore the possibilities for teaching the neural networks to generate completely

Inspired by the success reported in my last blog post [Link] about the open source docking program rDock [http://rdock.sourceforge.net], I decided to investigate the docking accuracy performance

Last time i tested the basics of the docking program rDock; Installation, basic setup and docking, as well as had a brief walkthrough about how to post-process
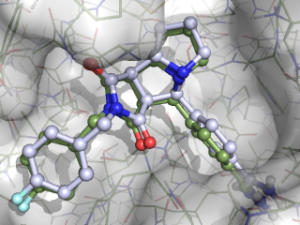
rDock is an open source docking program, trying to solve the same kind of scientific questions as Autodock Vina, which I have covered in a couple of
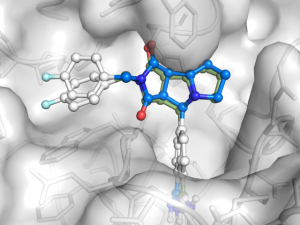
In the two previous blog posts Ligand docking with Smina and Never use re-docking for …, it was demonstrated how easy it is to dock a small ligand

Re-docking of ligands found in PDB files are often used as a fast evaluation of a docking program before working with designed or other ligands. However this
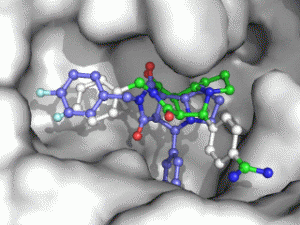
Moleculer Docking is a powerful technique for studying potential ligand-receptor interactions. It can be done with free tools. In this blog post I showcase the docking program
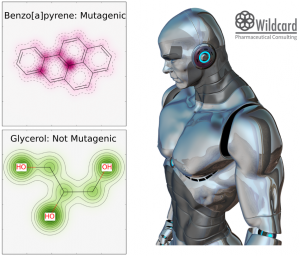
Toxic compounds are most often something that we try to avoid when designing novel pharmaceutical compounds, so it could be nice to get a prediction if a

Molecular mechanics treats molecules as ball and spring systems using classical mechanical physics. Molecules are modeled as ”balls on springs”, and the total conformational energy are described