I’d like to share a post about a project I’ve been involved in developing—Scikit-Mol. I believe it’s a noteworthy project deserving attention. It has already been featured


I’d like to share a post about a project I’ve been involved in developing—Scikit-Mol. I believe it’s a noteworthy project deserving attention. It has already been featured
We’ve known since 2016 that LSTM networks can be used to generate novel and valid SMILES strings of novel molecules after being trained on a dataset of
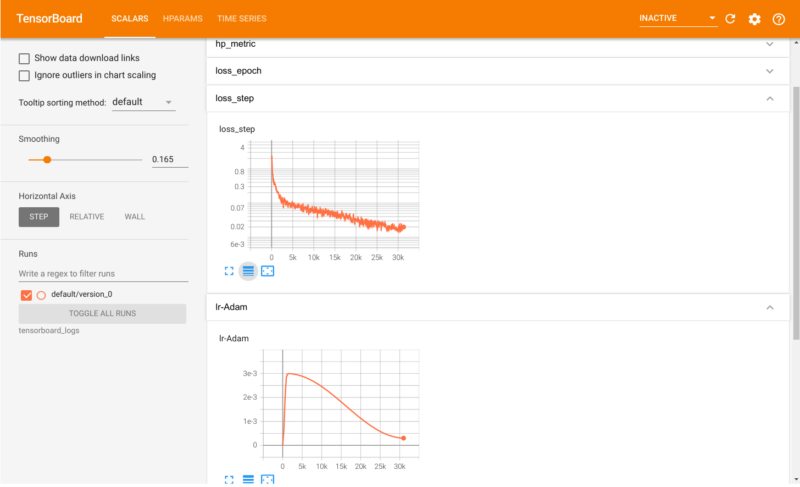
In the last blogpost I covered how LSTM-to-LSTM networks could be used to “translate” reactants into products of chemical reactions. Performance was however not very good of
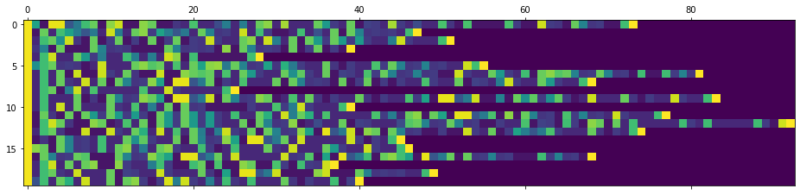
In this blogpost I’ll show how to predict chemical reactions with a sequence to sequence network based on LSTM cells. It’s the same principle as IBM’s RXN

I have been writing a lot about how to use SMILES together with deep learning architectures such as RNNs and LSTM networks to perform various cheminformatic and
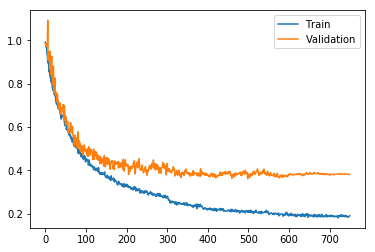
Last blog-post I showed how to use PyTorch to build a feed forward neural network model for molecular property prediction (QSAR: Quantitative structure-activity relationship). RDKit was used
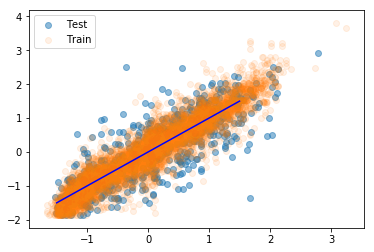
In my previous blogposts I’ve entirely been using Keras for my neural networks. Keras as a stand-alone is now no longer active developed, but are instead now
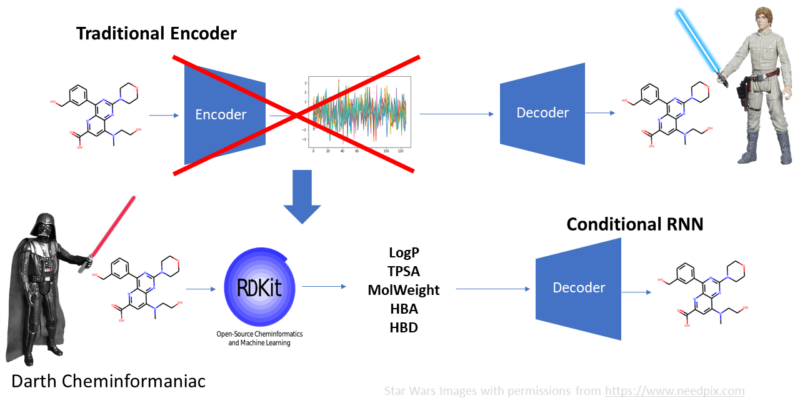
Long time ago in a GPU far-far away, the deep learning rebels are happy. They have created new ways of working with chemistry using deep learning technology
Not so long ago Greg Landrum published a blog post with an example of how the SVG rendering from RDKit in a jupyter notebook can be
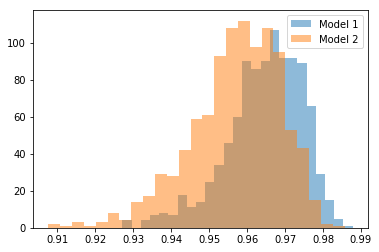
Some time ago I stumbled upon some work by Patrick Walters which shows that correlation coefficients have a rather large standard error when the sample sets sizes